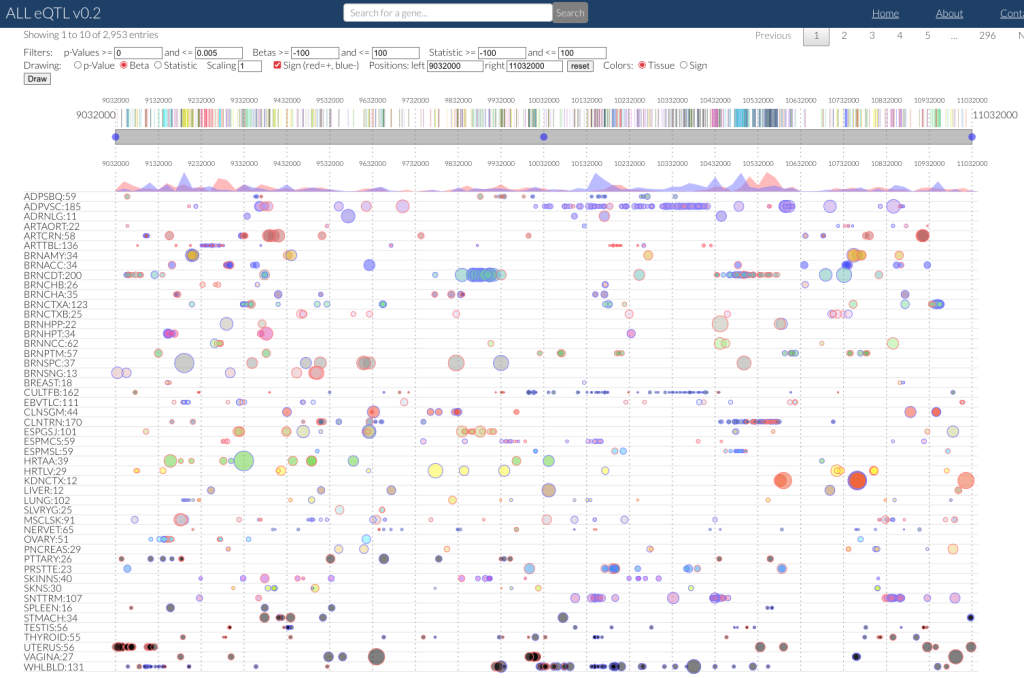
ALLeQTLs : cis-eQTLs from GTEx data (Farid Jacome’s Thesis)
CoGTex-2 : Views of Co-Expression from GTeX data (Maria Julia Teja)
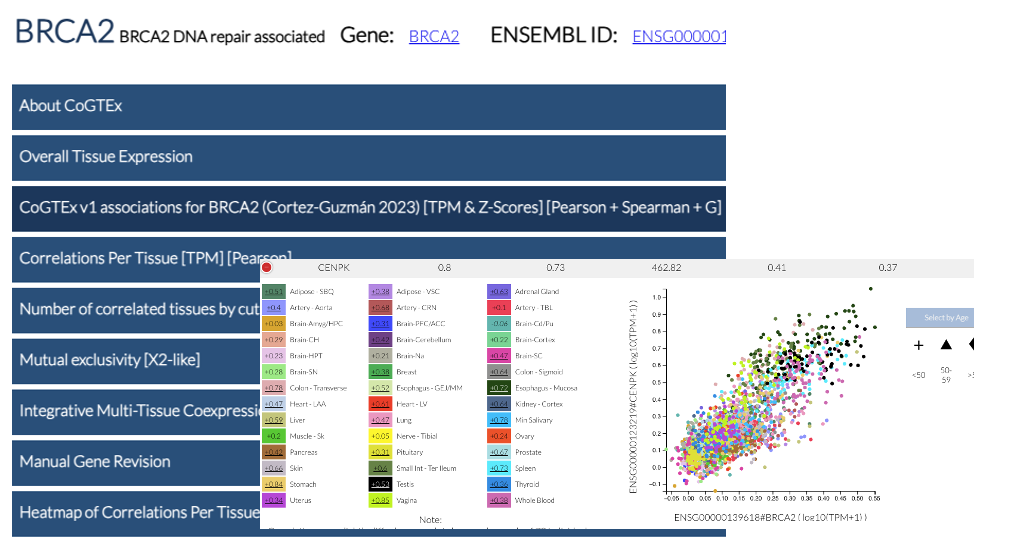
CoGTex: A Co-Expression Platform from GTeX data (Miguel Cortes Thesis). Paper on PLoS One

SurvHotspots: An informative database for survival analysis of cancer hotspots (Melissa Gonzalez Thesis). Paper on Cancers


PubTerm: A web-based annotation tool for PubMed articles, genes, or any other biomedical term. Paper on Databases (José Garcia-Pelaez).

SurvExpress: A web-based analysis of transcriptomic biomarkers in cancer. Paper on PLOS One (Raúl Aguirre-Gamboa Thesis).

HotspotsAnnotations: A web-based tool to annotate/visualize hotspots in cancer. Paper on Database.

SurvMicro: A web-based analysis of micro RNA expression biomarkers in cancer. (Server down). Paper on Bioinformatics. (Raúl Aguirre-Gamboa Thesis)
GALGO: Genetic Algorithms for Optimization. An R package for generic search optimizations. Paper on Bioinformatics.
See Galgo on CRAN R packages. Galgo Tutorial.
Galgo source code on GitHub

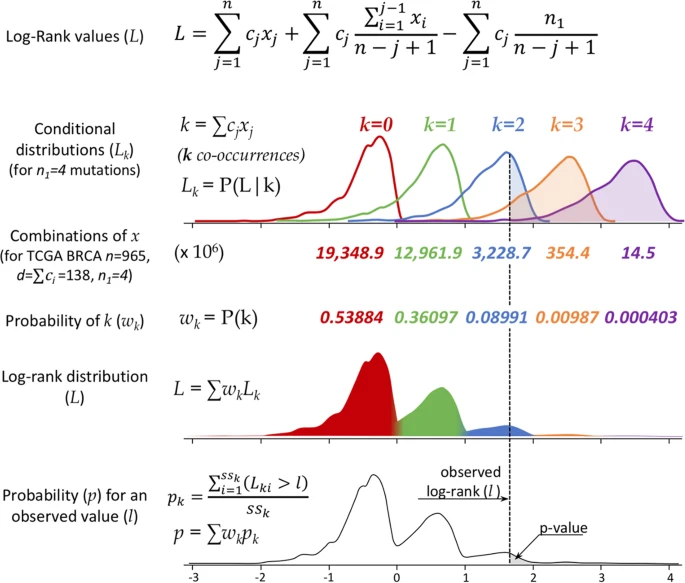
VALORATE: Velocity and Accuracy of the LOg-RAnk TEst. R package to quickly estimates the distribution of the log-rank especially for heavy unbalanced groups.
- CRAN package:
https://cran.r-project.org/web/packages/valorate/index.html - Beta/Early implementation:
https://github.com/vtrevino/valorate - Package Paper in Bioinformatics (PubMed):
https://pubmed.ncbi.nlm.nih.gov/28186225/ - Model Paper in Scientific Reports:
https://pubmed.ncbi.nlm.nih.gov/28240231/
GridMass: A MzMine add-on for peak detection (feature detection) in the analysis of metabolomics data. Paper on J on Mass Spectometry.
GridMass on MZMine 2, or MZMine 3.

PUBTERM CURATED GENE LISTS FROM OUR PUBLICATIONS
